Singular Value Decomposition
Contents
Live Notebook
You can run this notebook in a live session 
Singular Value Decomposition¶
This notebook introduces the da.linalg.svd algorithms for the Singular Value Decomposition
Start Dask Client for Dashboard¶
Starting the Dask Client is optional. It will provide a dashboard which is useful to gain insight on the computation.
The link to the dashboard will become visible when you create the client below. We recommend having it open on one side of your screen while using your notebook on the other side. This can take some effort to arrange your windows, but seeing them both at the same is very useful when learning.
[1]:
from dask.distributed import Client, progress
client = Client(processes=False, threads_per_worker=4,
n_workers=1, memory_limit='2GB')
client
[1]:
Client
Client-8ad1cc86-0de1-11ed-a4f6-000d3a8f7959
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://10.1.1.64:8787/status |
Cluster Info
LocalCluster
9a88c089
| Dashboard: http://10.1.1.64:8787/status | Workers: 1 |
| Total threads: 4 | Total memory: 1.86 GiB |
| Status: running | Using processes: False |
Scheduler Info
Scheduler
Scheduler-ad8714da-ab0d-4c50-a4cb-ec68028ac54d
| Comm: inproc://10.1.1.64/9462/1 | Workers: 1 |
| Dashboard: http://10.1.1.64:8787/status | Total threads: 4 |
| Started: Just now | Total memory: 1.86 GiB |
Workers
Worker: 0
| Comm: inproc://10.1.1.64/9462/4 | Total threads: 4 |
| Dashboard: http://10.1.1.64:39957/status | Memory: 1.86 GiB |
| Nanny: None | |
| Local directory: /home/runner/work/dask-examples/dask-examples/machine-learning/dask-worker-space/worker-we6_wppm | |
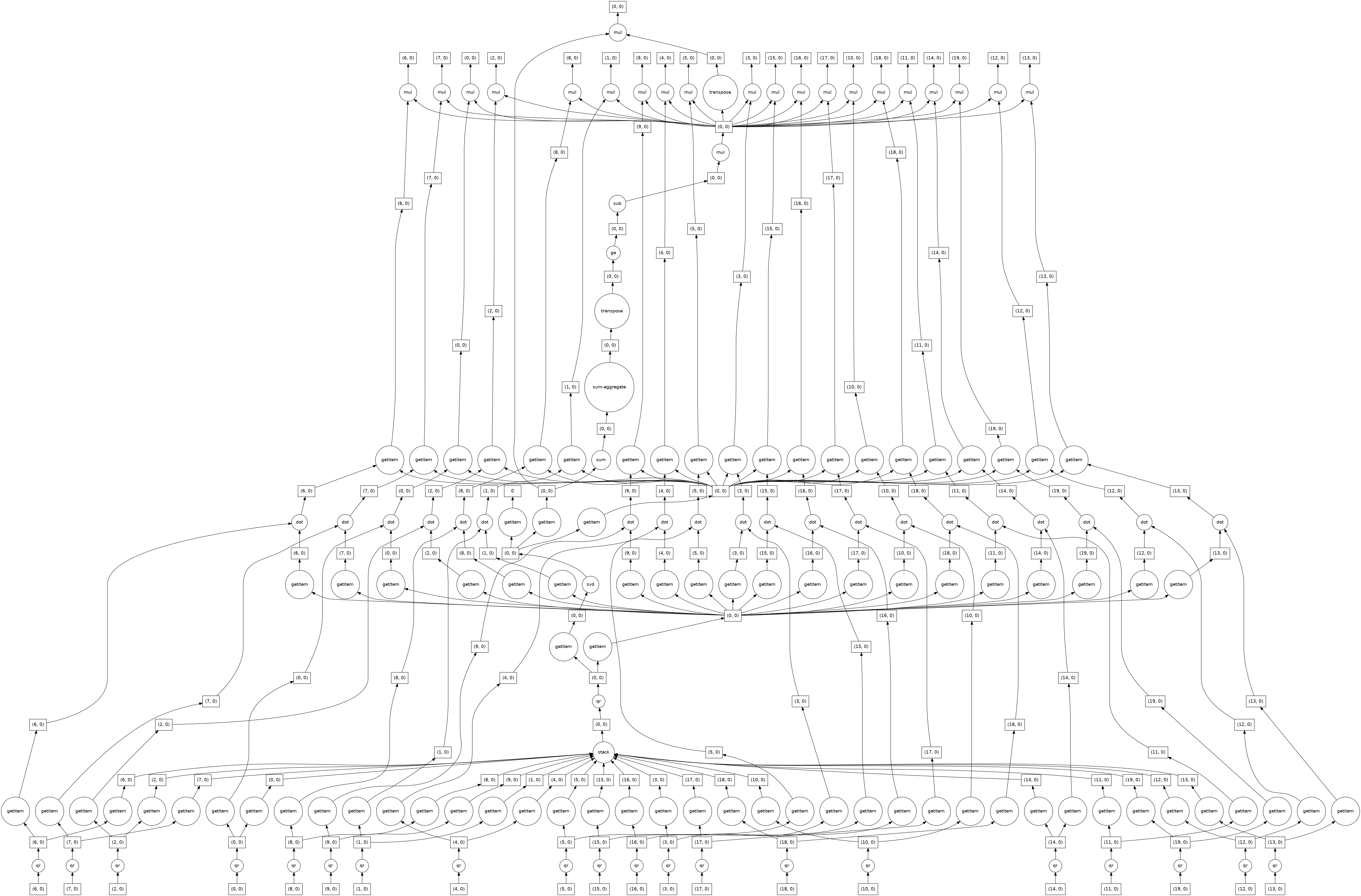
Compute SVD of Tall-and-Skinny Matrix¶
For many applications the provided matrix has many more rows than columns. In this case a specialized algorithm can be used.
[2]:
import dask.array as da
X = da.random.random((200000, 100), chunks=(10000, 100)).persist()
[3]:
import dask
u, s, v = da.linalg.svd(X)
dask.visualize(u, s, v)
[3]:
[4]:
v.compute()
[4]:
array([[ 0.09994831, 0.10007229, 0.09997617, ..., 0.09995264,
0.09995591, 0.09984163],
[ 0.05585195, -0.06184545, -0.04747733, ..., -0.06275421,
-0.2061527 , 0.18218227],
[ 0.01277435, 0.00484692, -0.04387551, ..., 0.00444605,
0.12143905, -0.06438531],
...,
[ 0.02933106, 0.00834248, 0.0103009 , ..., -0.06069817,
0.01291796, 0.12832988],
[ 0.0901224 , -0.00492353, -0.00470015, ..., 0.14196305,
-0.09734339, -0.05803211],
[ 0.16619815, 0.14906927, -0.18081339, ..., -0.1346468 ,
0.12524437, 0.01322112]])
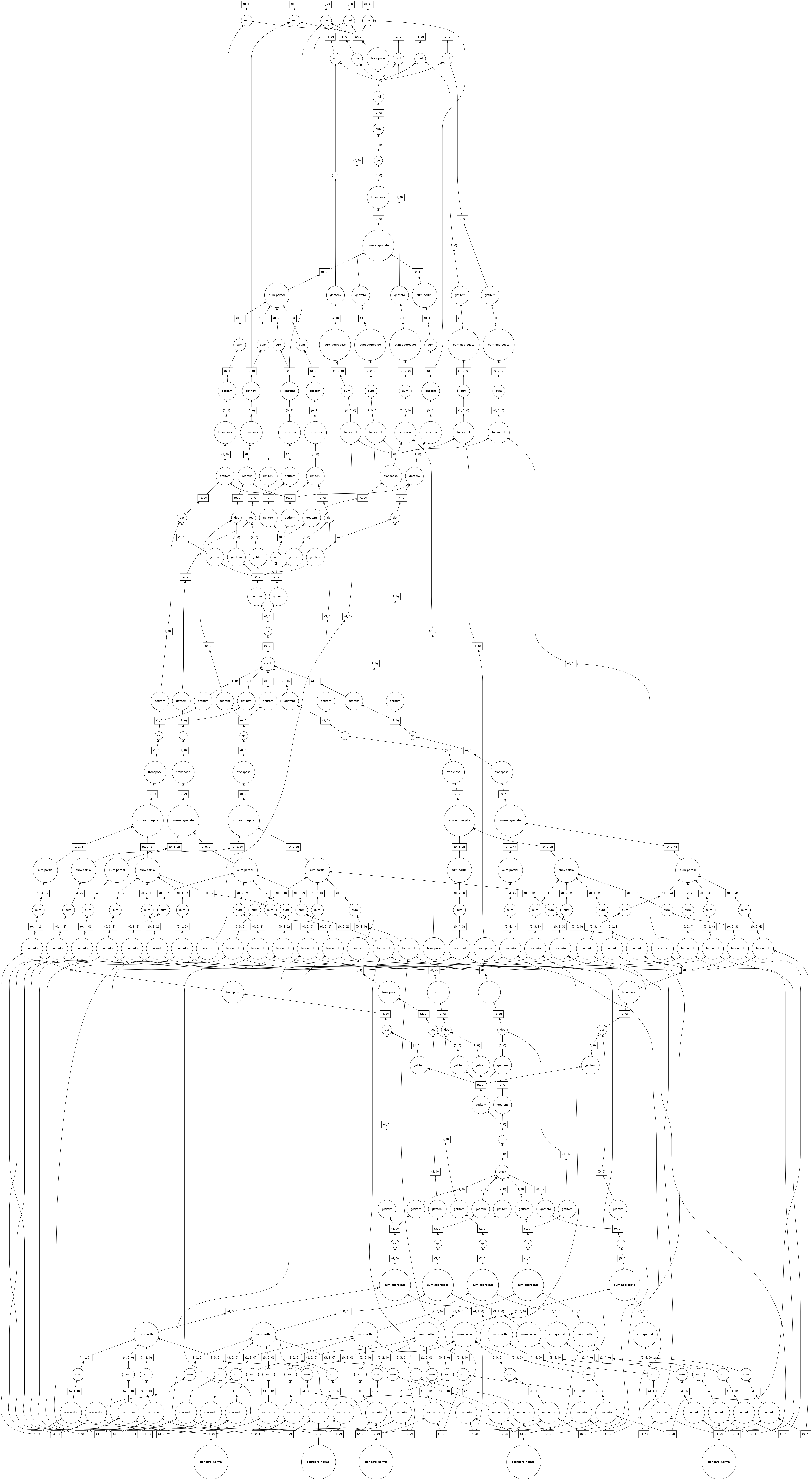
Compute SVD of General Non-Skinny Matrix with Approximate algorithm¶
When there are also many chunks in columns then we use an approximate randomized algorithm to collect only a few of the singular values and vectors.
[5]:
import dask.array as da
X = da.random.random((10000, 10000), chunks=(2000, 2000)).persist()
[6]:
import dask
u, s, v = da.linalg.svd_compressed(X, k=5)
dask.visualize(u, s, v)
[6]:
[7]:
v.compute()
[7]:
array([[ 0.00997573, 0.01011564, 0.00997098, ..., 0.00998413,
0.00994515, 0.0099736 ],
[-0.00048293, 0.00247921, -0.00527027, ..., -0.00606932,
-0.01272348, 0.00818248],
[ 0.00145378, -0.00278148, 0.01078658, ..., -0.00428512,
-0.00213905, -0.00738961],
[-0.00984979, -0.00230993, -0.00277437, ..., 0.0056367 ,
-0.00199535, -0.01409744],
[-0.00210629, -0.00320545, -0.00190336, ..., 0.01871436,
-0.01494592, -0.00274385]])